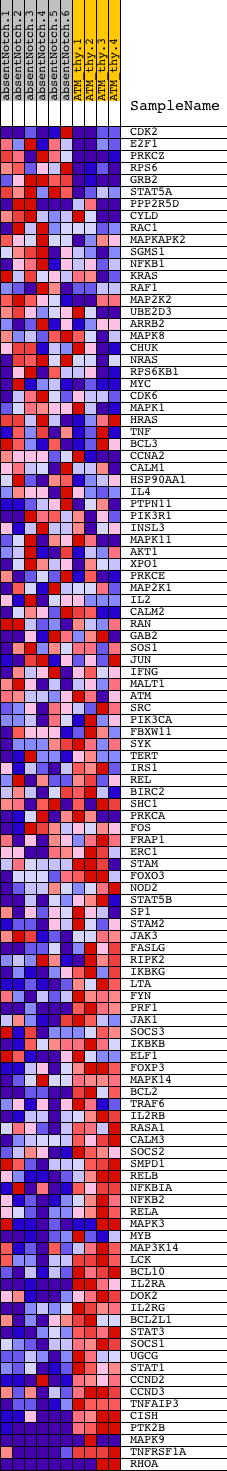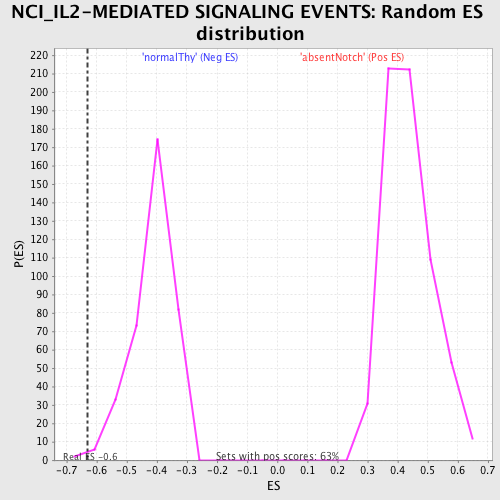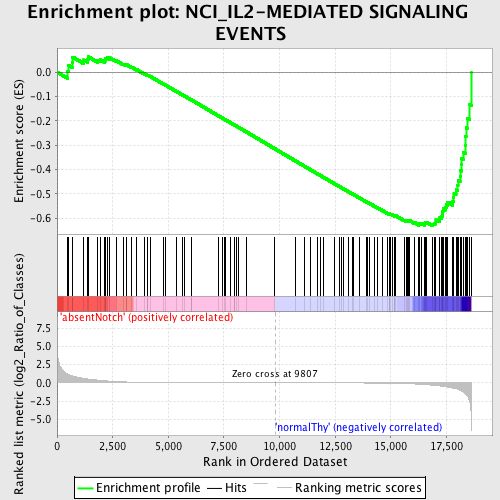
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | NCI_IL2-MEDIATED SIGNALING EVENTS |
| Enrichment Score (ES) | -0.63146526 |
| Normalized Enrichment Score (NES) | -1.5365051 |
| Nominal p-value | 0.010810811 |
| FDR q-value | 0.22672687 |
| FWER p-Value | 0.941 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CDK2 | 130484 2260301 4010088 5050110 | 481 | 1.197 | 0.0022 | No | ||
| 2 | E2F1 | 5360093 | 513 | 1.141 | 0.0275 | No | ||
| 3 | PRKCZ | 3780279 | 677 | 0.945 | 0.0410 | No | ||
| 4 | RPS6 | 2060168 | 695 | 0.929 | 0.0620 | No | ||
| 5 | GRB2 | 6650398 | 1188 | 0.612 | 0.0499 | No | ||
| 6 | STAT5A | 2680458 | 1375 | 0.523 | 0.0522 | No | ||
| 7 | PPP2R5D | 4010156 380408 5550112 5670162 | 1406 | 0.512 | 0.0626 | No | ||
| 8 | CYLD | 6590079 | 1812 | 0.370 | 0.0495 | No | ||
| 9 | RAC1 | 4810687 | 1942 | 0.331 | 0.0503 | No | ||
| 10 | MAPKAPK2 | 2850500 | 2120 | 0.284 | 0.0475 | No | ||
| 11 | SGMS1 | 4780195 | 2157 | 0.277 | 0.0520 | No | ||
| 12 | NFKB1 | 5420358 | 2177 | 0.270 | 0.0574 | No | ||
| 13 | KRAS | 2060170 | 2245 | 0.255 | 0.0598 | No | ||
| 14 | RAF1 | 1770600 | 2375 | 0.227 | 0.0582 | No | ||
| 15 | MAP2K2 | 4590601 | 2672 | 0.174 | 0.0463 | No | ||
| 16 | UBE2D3 | 3190452 | 2980 | 0.131 | 0.0328 | No | ||
| 17 | ARRB2 | 2060441 | 3118 | 0.115 | 0.0281 | No | ||
| 18 | MAPK8 | 2640195 | 3135 | 0.114 | 0.0299 | No | ||
| 19 | CHUK | 7050736 | 3349 | 0.094 | 0.0206 | No | ||
| 20 | NRAS | 6900577 | 3584 | 0.077 | 0.0098 | No | ||
| 21 | RPS6KB1 | 1450427 5080110 6200563 | 3919 | 0.057 | -0.0069 | No | ||
| 22 | MYC | 380541 4670170 | 4075 | 0.051 | -0.0141 | No | ||
| 23 | CDK6 | 4920253 | 4077 | 0.051 | -0.0129 | No | ||
| 24 | MAPK1 | 3190193 6200253 | 4185 | 0.046 | -0.0176 | No | ||
| 25 | HRAS | 1980551 | 4798 | 0.029 | -0.0500 | No | ||
| 26 | TNF | 6650603 | 4887 | 0.028 | -0.0541 | No | ||
| 27 | BCL3 | 3990440 | 5383 | 0.020 | -0.0804 | No | ||
| 28 | CCNA2 | 5290075 | 5631 | 0.018 | -0.0933 | No | ||
| 29 | CALM1 | 380128 | 5743 | 0.017 | -0.0989 | No | ||
| 30 | HSP90AA1 | 4560041 5220133 2120722 | 6024 | 0.015 | -0.1137 | No | ||
| 31 | IL4 | 6020537 | 6051 | 0.014 | -0.1148 | No | ||
| 32 | PTPN11 | 2230100 2470180 6100528 | 7257 | 0.008 | -0.1797 | No | ||
| 33 | PIK3R1 | 4730671 | 7437 | 0.007 | -0.1892 | No | ||
| 34 | INSL3 | 4150092 | 7532 | 0.007 | -0.1941 | No | ||
| 35 | MAPK11 | 130452 3120440 3610465 | 7589 | 0.007 | -0.1970 | No | ||
| 36 | AKT1 | 5290746 | 7797 | 0.006 | -0.2080 | No | ||
| 37 | XPO1 | 540707 | 7981 | 0.005 | -0.2178 | No | ||
| 38 | PRKCE | 5700053 | 7983 | 0.005 | -0.2177 | No | ||
| 39 | MAP2K1 | 840739 | 8060 | 0.005 | -0.2217 | No | ||
| 40 | IL2 | 1770725 | 8136 | 0.005 | -0.2256 | No | ||
| 41 | CALM2 | 6620463 | 8534 | 0.004 | -0.2470 | No | ||
| 42 | RAN | 2260446 4590647 | 9782 | 0.000 | -0.3144 | No | ||
| 43 | GAB2 | 1410280 2340520 4280040 | 10726 | -0.003 | -0.3653 | No | ||
| 44 | SOS1 | 7050338 | 11108 | -0.004 | -0.3858 | No | ||
| 45 | JUN | 840170 | 11404 | -0.005 | -0.4016 | No | ||
| 46 | IFNG | 5670592 | 11695 | -0.006 | -0.4172 | No | ||
| 47 | MALT1 | 4670292 | 11831 | -0.006 | -0.4243 | No | ||
| 48 | ATM | 3610110 4050524 | 11982 | -0.007 | -0.4322 | No | ||
| 49 | SRC | 580132 | 12478 | -0.009 | -0.4588 | No | ||
| 50 | PIK3CA | 6220129 | 12684 | -0.010 | -0.4696 | No | ||
| 51 | FBXW11 | 6450632 | 12795 | -0.011 | -0.4753 | No | ||
| 52 | SYK | 6940133 | 12885 | -0.012 | -0.4798 | No | ||
| 53 | TERT | 6660075 | 13081 | -0.013 | -0.4900 | No | ||
| 54 | IRS1 | 1190204 | 13277 | -0.014 | -0.5002 | No | ||
| 55 | REL | 360707 | 13310 | -0.015 | -0.5016 | No | ||
| 56 | BIRC2 | 2940435 4730020 | 13581 | -0.017 | -0.5158 | No | ||
| 57 | SHC1 | 2900731 3170504 6520537 | 13907 | -0.021 | -0.5329 | No | ||
| 58 | PRKCA | 6400551 | 13942 | -0.022 | -0.5342 | No | ||
| 59 | FOS | 1850315 | 14022 | -0.023 | -0.5379 | No | ||
| 60 | FRAP1 | 2850037 6660010 | 14253 | -0.027 | -0.5497 | No | ||
| 61 | ERC1 | 1580170 2630717 6110762 6840280 | 14388 | -0.030 | -0.5562 | No | ||
| 62 | STAM | 3850008 4760441 | 14646 | -0.036 | -0.5693 | No | ||
| 63 | FOXO3 | 2510484 4480451 | 14837 | -0.044 | -0.5785 | No | ||
| 64 | NOD2 | 2510050 | 14922 | -0.048 | -0.5819 | No | ||
| 65 | STAT5B | 6200026 | 14943 | -0.049 | -0.5818 | No | ||
| 66 | SP1 | 6590017 | 14989 | -0.051 | -0.5831 | No | ||
| 67 | STAM2 | 730731 2120377 | 15071 | -0.056 | -0.5861 | No | ||
| 68 | JAK3 | 70347 3290008 | 15166 | -0.062 | -0.5897 | No | ||
| 69 | FASLG | 2810044 | 15181 | -0.063 | -0.5890 | No | ||
| 70 | RIPK2 | 5050072 6290632 | 15223 | -0.066 | -0.5896 | No | ||
| 71 | IKBKG | 3450092 3840377 6590592 | 15620 | -0.102 | -0.6086 | No | ||
| 72 | LTA | 1740088 | 15702 | -0.110 | -0.6104 | No | ||
| 73 | FYN | 2100468 4760520 4850687 | 15761 | -0.116 | -0.6108 | No | ||
| 74 | PRF1 | 6660309 | 15795 | -0.120 | -0.6097 | No | ||
| 75 | JAK1 | 5910746 | 15852 | -0.127 | -0.6098 | No | ||
| 76 | SOCS3 | 5550563 | 16074 | -0.155 | -0.6181 | No | ||
| 77 | IKBKB | 6840072 | 16263 | -0.180 | -0.6240 | Yes | ||
| 78 | ELF1 | 4780450 | 16294 | -0.185 | -0.6212 | Yes | ||
| 79 | FOXP3 | 940707 1850692 | 16393 | -0.203 | -0.6217 | Yes | ||
| 80 | MAPK14 | 5290731 | 16518 | -0.228 | -0.6231 | Yes | ||
| 81 | BCL2 | 730132 1570736 2470138 3800044 4810037 5690068 5860504 6650164 | 16536 | -0.234 | -0.6185 | Yes | ||
| 82 | TRAF6 | 4810292 6200132 | 16620 | -0.253 | -0.6170 | Yes | ||
| 83 | IL2RB | 4730072 | 16889 | -0.317 | -0.6240 | Yes | ||
| 84 | RASA1 | 1240315 | 16954 | -0.335 | -0.6195 | Yes | ||
| 85 | CALM3 | 3390288 | 17028 | -0.355 | -0.6151 | Yes | ||
| 86 | SOCS2 | 4760692 | 17029 | -0.355 | -0.6067 | Yes | ||
| 87 | SMPD1 | 1410309 5550717 | 17184 | -0.408 | -0.6054 | Yes | ||
| 88 | RELB | 1400048 | 17205 | -0.414 | -0.5967 | Yes | ||
| 89 | NFKBIA | 1570152 | 17293 | -0.450 | -0.5908 | Yes | ||
| 90 | NFKB2 | 2320670 | 17315 | -0.460 | -0.5811 | Yes | ||
| 91 | RELA | 3830075 | 17328 | -0.467 | -0.5707 | Yes | ||
| 92 | MAPK3 | 580161 4780035 | 17348 | -0.472 | -0.5606 | Yes | ||
| 93 | MYB | 1660494 5860451 6130706 | 17450 | -0.518 | -0.5538 | Yes | ||
| 94 | MAP3K14 | 5890435 | 17492 | -0.536 | -0.5434 | Yes | ||
| 95 | LCK | 3360142 | 17548 | -0.561 | -0.5331 | Yes | ||
| 96 | BCL10 | 2360397 | 17788 | -0.705 | -0.5294 | Yes | ||
| 97 | IL2RA | 6620450 | 17824 | -0.729 | -0.5140 | Yes | ||
| 98 | DOK2 | 5340273 | 17838 | -0.737 | -0.4974 | Yes | ||
| 99 | IL2RG | 4120273 | 17931 | -0.809 | -0.4832 | Yes | ||
| 100 | BCL2L1 | 1580452 4200152 5420484 | 18009 | -0.884 | -0.4665 | Yes | ||
| 101 | STAT3 | 460040 3710341 | 18021 | -0.893 | -0.4461 | Yes | ||
| 102 | SOCS1 | 730139 | 18113 | -1.004 | -0.4273 | Yes | ||
| 103 | UGCG | 3420053 | 18140 | -1.039 | -0.4042 | Yes | ||
| 104 | STAT1 | 6510204 6590553 | 18176 | -1.089 | -0.3803 | Yes | ||
| 105 | CCND2 | 5340167 | 18184 | -1.101 | -0.3547 | Yes | ||
| 106 | CCND3 | 380528 730500 | 18253 | -1.212 | -0.3298 | Yes | ||
| 107 | TNFAIP3 | 2900142 | 18358 | -1.497 | -0.3001 | Yes | ||
| 108 | CISH | 840315 | 18365 | -1.521 | -0.2645 | Yes | ||
| 109 | PTK2B | 4730411 | 18382 | -1.574 | -0.2282 | Yes | ||
| 110 | MAPK9 | 2060273 3780209 4070397 | 18430 | -1.744 | -0.1896 | Yes | ||
| 111 | TNFRSF1A | 1090390 6520735 | 18550 | -2.617 | -0.1343 | Yes | ||
| 112 | RHOA | 580142 5900131 5340450 | 18613 | -5.837 | 0.0002 | Yes |